Example single fit¶
Fitting method
Read data
Select and create model
Fit the data with model
Save plots and log
Quick fit¶
import src.binary_sed_fitting as bsf
################################################################################
name = 'WOCS2002'
file_name = 'data/extinction_corrected_flux_files/%s.csv'%name
data = bsf.load_data(file_name, mode='csv')
distance = 831. # pc
e_distance = 11. # pc
################################################################################
model_name = 'kurucz'
limits = {'Te' : [3500, 9000],
'logg' : [ 3, 5],
'MH' : [ 0.0, 0.0],
'alpha': [ 0.0, 0.0]}
model = bsf.Model(model_name, limits=limits)
################################################################################
star = bsf.Star(name=name,
distance=distance,
e_distance=e_distance,
data=data,
model=model)
################################################################################
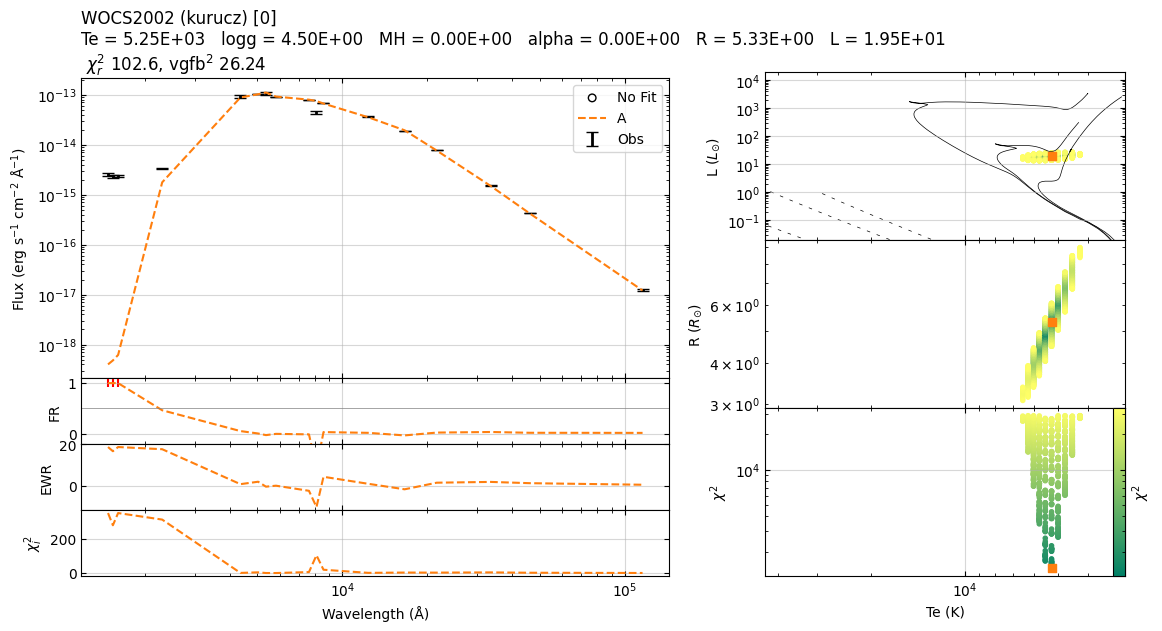
star.fit_chi2()
star.plot()
22:39:52 ----- WARNING ----- estimate_runtime
Calculating chi2: ETA ~ 0 s
Selecting model¶
Supported models: Kurucz, Koester and Kurucz_UVBLUE
import src.binary_sed_fitting as bsf
################################################################################
model_name = 'kurucz'
limits = {'Te' : [3500, 9000],
'logg' : [ 3, 5],
'MH' : [ 0.0, 0.0],
'alpha': [ 0.0, 0.0]}
model = bsf.Model(model_name, limits=limits)
print(model_name, '\n\n\n' ,model.da)
################################################################################
model_name = 'kurucz_uvblue'
limits = {'Te' : [3500, 9000],
'logg' : [ 3, 5],
'MH' : [ 0.0, 0.0]}
model = bsf.Model(model_name, limits=limits)
print(model_name, '\n\n\n' ,model.da)
################################################################################
model_name = 'koester'
limits = {'Te' : [5000, 80000],
'logg' : [ 6.5, 9.5]}
model = bsf.Model(model_name, limits=limits)
print(model_name, '\n\n\n' ,model.da)
kurucz
<xarray.DataArray (FilterID: 8162, Te: 23, logg: 5, MH: 1, alpha: 1)> Size: 8MB
[938630 values with dtype=float64]
Coordinates:
* FilterID (FilterID) <U38 1MB 'Swift/UVOT.UVM2_fil' ... 'QUIJOTE/MFI.11GH...
* Te (Te) int32 92B 3500 3750 4000 4250 4500 ... 8250 8500 8750 9000
* logg (logg) float64 40B 3.0 3.5 4.0 4.5 5.0
* MH (MH) float64 8B 0.0
* alpha (alpha) float64 8B 0.0
Attributes:
Wavelengths: [1.19128077e+00 1.24254209e+00 1.48134125e+02 ... 3.8683349...
unit: erg/s/cm2/A
long_name: Flux
kurucz_uvblue
<xarray.DataArray (FilterID: 8162, Te: 12, logg: 5, MH: 1)> Size: 4MB
[489720 values with dtype=float64]
Coordinates:
* FilterID (FilterID) <U38 1MB 'Swift/UVOT.UVM2_fil' ... 'QUIJOTE/MFI.11GH...
* Te (Te) int32 48B 3500 4000 4500 5000 5500 ... 7500 8000 8500 9000
* logg (logg) float64 40B 3.0 3.5 4.0 4.5 5.0
* MH (MH) float64 8B 0.0
alpha float64 8B ...
Attributes:
Wavelengths: [1.19128077e+00 1.24254209e+00 1.48134125e+02 ... 3.8683349...
unit: erg/s/cm2/A
long_name: Flux
koester
<xarray.DataArray (FilterID: 8162, Te: 82, logg: 13)> Size: 70MB
[8700692 values with dtype=float64]
Coordinates:
* FilterID (FilterID) <U38 1MB 'IUE/IUE.1250-1300' ... 'QUIJOTE/MFI.11GHz_H3'
* Te (Te) int32 328B 5000 5250 5500 5750 ... 50000 60000 70000 80000
* logg (logg) float64 104B 6.5 6.75 7.0 7.25 7.5 ... 8.75 9.0 9.25 9.5
Attributes:
Wavelengths: [1.28470318e+03 1.35336435e+03 1.35623969e+03 ... 3.8683349...
unit: erg/s/cm2/A
long_name: Flux
Recommended fitting routine¶
Starname: Name of the stardistance,e_distance: Distance and it’s error in pcfilters_to_drop: Filters to be removed from the fit (due to bad data, bad fit, saturation…)wavelength_range: Wavelength range to be fitted in Angstromdata: DataFrame with the photometric fluxmodel: Model object of required model and parameter limitsr_limits: 2 optionsblackbody: Radius range automatically calculated from blackbody fit[r_min,r_max]: Radius is varied between the r_min and r_max given in solar radii
run_name: A string name for tracking different fits
fit_chi2Calculates chi2 for the data and model.
refitif
True: the fit is redone.If
False: If a previous fit is available, it is read. Otherwise a fresh fit is made.
fit_noisy_chi2Fits chi2 by adding noise to the data. Useful for getting statistical errors.
refit: ifTruethe fit is redone. IfFalse, the previous fit is read.
plotadd_noisy_seds: Whether to plot the noisy SEDs.folder: Folder for saving the plotFR_cutoff: Fractional residual cutoff for indicating excess flux
plot_publicPlots a smaller plot with SED, FR and EWR
save_summarySaves the fit parameters in a logfile
Example 1¶
import src.binary_sed_fitting as bsf
import warnings
warnings.filterwarnings("ignore")
bsf.console.setLevel(bsf.logging.INFO)
################################################################################
name = 'WOCS2002'
file_name = 'data/extinction_corrected_flux_files/%s.csv'%name
data = bsf.load_data(file_name, mode='csv')
distance = 831. # pc
e_distance = 11. # pc
refit = False
run_name = '0'
################################################################################
model_name = 'kurucz'
limits = {'Te' : [3500, 9000],
'logg' : [ 3, 5],
'MH' : [ 0.0, 0.0],
'alpha': [ 0.0, 0.0]}
model = bsf.Model(model_name, limits=limits)
################################################################################
star = bsf.Star(name=name,
distance=distance,
e_distance=e_distance,
filters_to_drop=['KPNO/Mosaic.I'],
wavelength_range=[3000, 1_000_000_000],
data=data,
model=model,
r_limits='blackbody',
run_name=run_name)
################################################################################
star.fit_chi2(refit=refit)
star.fit_noisy_chi2(refit=refit)
################################################################################
star.plot(add_noisy_seds=False,
folder='plots/',
FR_cutoff=0.5)
star.plot_public(add_noisy_seds=False,
folder='plots/',
FR_cutoff=0.5)
################################################################################
star.save_summary()
22:39:54 ----- INFO ----- __init__
==========================================================
----------------------------------------------------------
------------ WOCS2002 A ------------
----------------------------------------------------------
==========================================================
22:39:54 ----- INFO ----- drop_filters
Fitted Not fitted
wavelength
1481.000000 Astrosat/UVIT.F148W
1541.000000 Astrosat/UVIT.F154W
1608.000000 Astrosat/UVIT.F169M
2303.366368 GALEX/GALEX.NUV
4357.276538 KPNO/Mosaic.B
5035.750275 GAIA/GAIA3.Gbp
5366.240786 KPNO/Mosaic.V
5822.388714 GAIA/GAIA3.G
7619.959993 GAIA/GAIA3.Grp
8101.609574 KPNO/Mosaic.I
8578.159519 GAIA/GAIA3.Grvs
12350.000000 2MASS/2MASS.J
16620.000000 2MASS/2MASS.H
21590.000000 2MASS/2MASS.Ks
33526.000000 WISE/WISE.W1
46028.000000 WISE/WISE.W2
115608.000000 WISE/WISE.W3
22:39:54 ----- INFO ----- drop_filters
Filters: used/all = 12/17
22:39:54 ----- INFO ----- blackbody
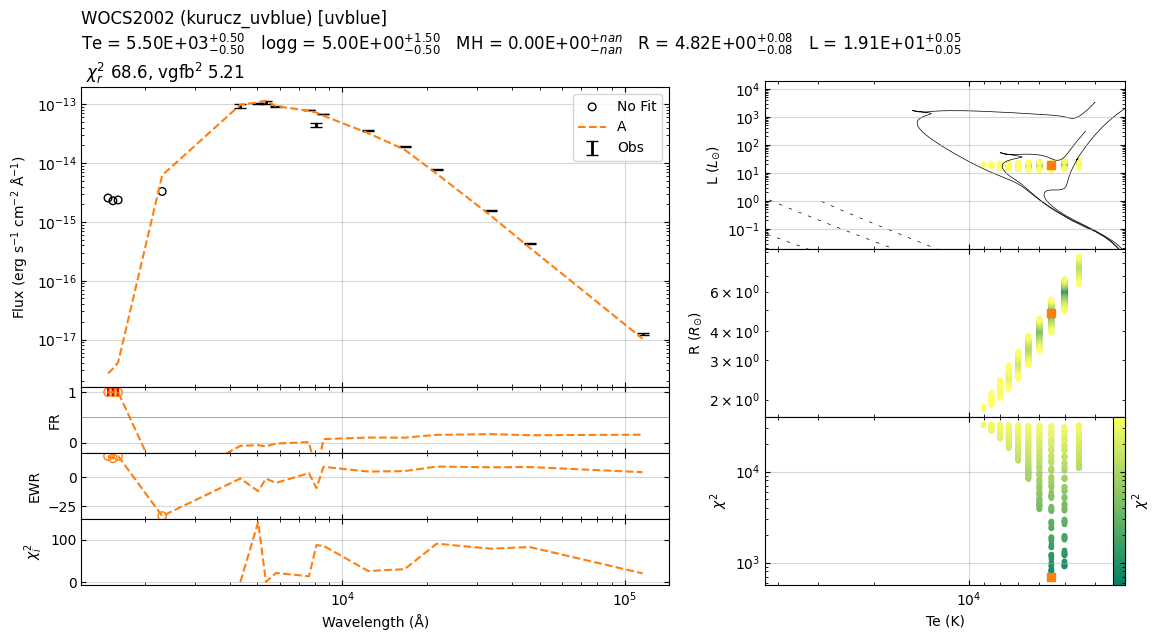
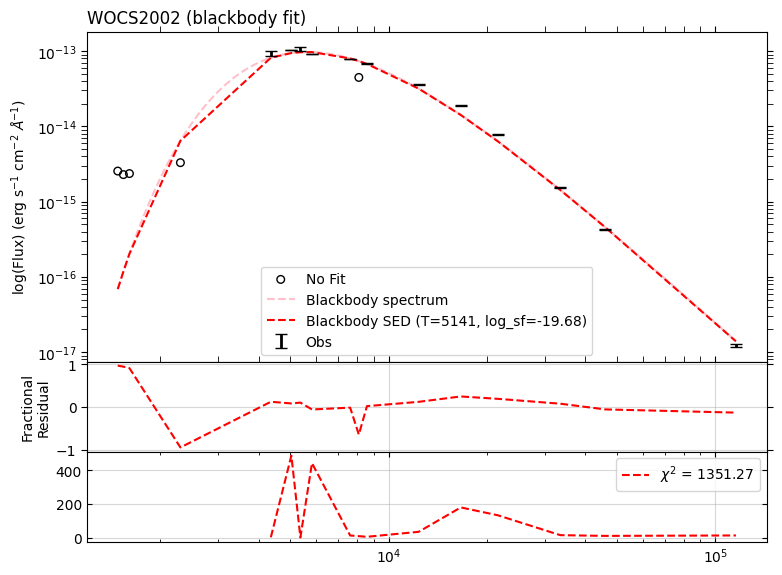
Fit parameters: T=5141 K, log_sf=-19.68
22:39:54 ----- WARNING ----- calculate_chi2
Give "refit=True" if you want to rerun the fitting process.
22:39:54 ----- INFO ----- calculate_chi2
Te logg MH alpha sf chi2 R L
0 5250 4.5 0.0 0.0 2.092353e-20 1436.600470 5.331470 19.454716
1 5250 4.0 0.0 0.0 2.092353e-20 1437.936819 5.331470 19.454716
2 5250 5.0 0.0 0.0 2.092353e-20 1465.574398 5.331470 19.454716
3 5250 3.5 0.0 0.0 2.092353e-20 1475.514792 5.331470 19.454716
4 5250 4.5 0.0 0.0 2.141090e-20 1557.722256 5.393206 19.907874
22:39:54 ----- INFO ----- get_parameters_from_chi2_minimization
Te 5250
logg 4.5
MH 0.0
alpha 0.0
sf 2.0923525610805822e-20
chi2 1436.6004696876382
R 5.331470347607279
L 19.454715675310812
vgf2 1409.3254983079803
vgfb2 367.3732364927856
22:39:54 ----- WARNING ----- get_parameters_from_chi2_minimization
Based on chi2, I recommend removal of following filters: ['GAIA/GAIA3.Grvs']; chi2=[19.2204403]
22:39:54 ----- WARNING ----- calculate_noisy_chi2
Give "refit=True" if you want to rerun the fitting process.
22:39:54 ----- INFO ----- calculate_noisy_chi2
Te logg MH alpha sf chi2 R L
0 5250 4.5 0.0 0.0 2.092353e-20 1436.600470 5.33147 19.454716
1 5250 4.5 0.0 0.0 2.092353e-20 1448.373499 5.33147 19.454716
2 5250 4.0 0.0 0.0 2.092353e-20 1365.291238 5.33147 19.454716
3 5250 4.5 0.0 0.0 2.092353e-20 1384.661354 5.33147 19.454716
4 5250 4.5 0.0 0.0 2.092353e-20 1519.437073 5.33147 19.454716
22:39:54 ----- INFO ----- get_parameters_from_noisy_chi2_minimization
Te 5250(-250,+250)
logg 4.5(-0.5,+0.5)
MH 0.0(-nan,+nan)
alpha 0.0(-nan,+nan)
sf 2.0923525610805822e-20(-4.762775903042835e-22,+4.8737152053873975e-22)
R 5.331470347607279(-0.0933009594847496,+0.09376472807053667)
L 19.454715675310812(-0.5150466243764595,+0.5150466243764595)
22:39:55 ----- INFO ----- save_summary
Saving log in data/log_single_fitting.csv
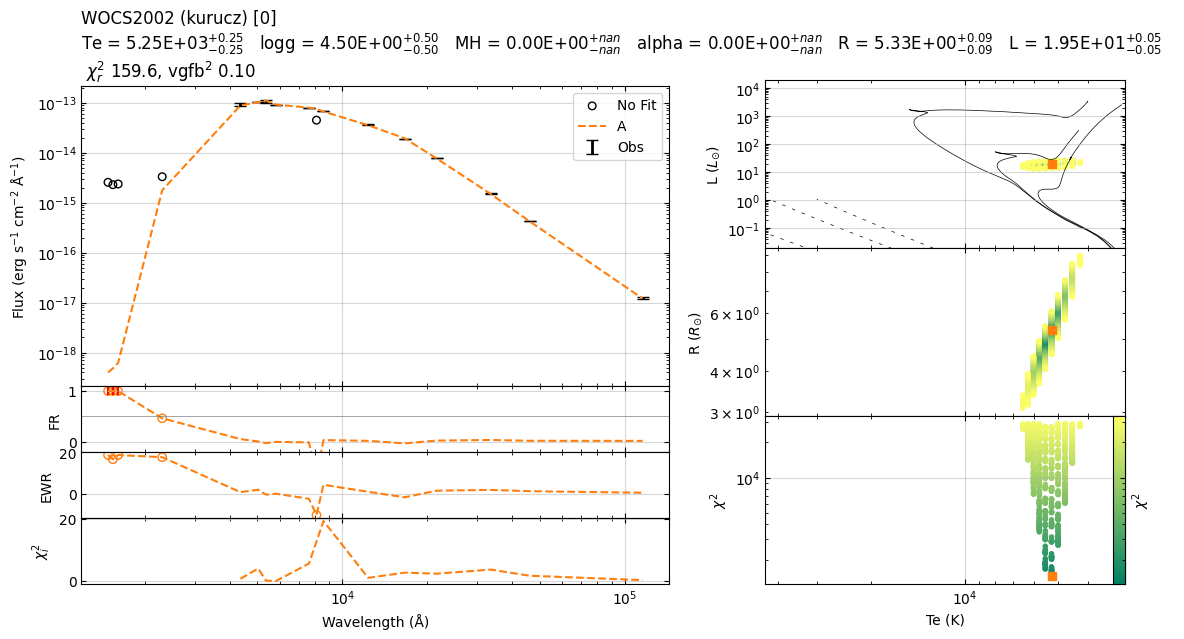
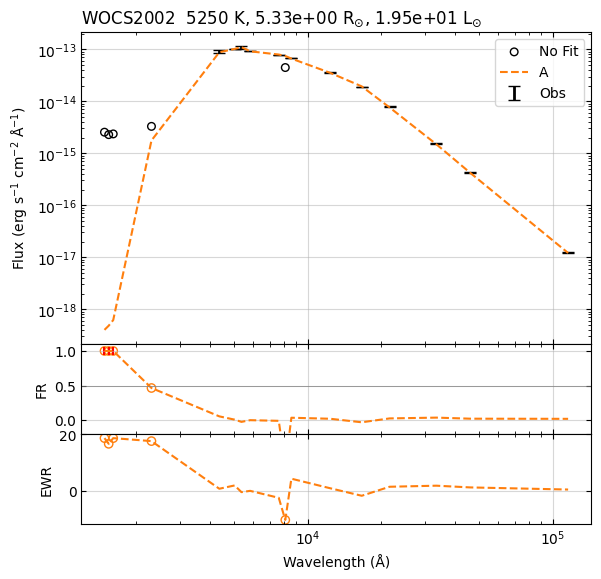
Example 2¶
import src.binary_sed_fitting as bsf
import warnings
warnings.filterwarnings("ignore")
bsf.console.setLevel(bsf.logging.WARNING)
################################################################################
name = 'Y1168'
file_name = 'data/extinction_corrected_flux_files/%s.csv'%name
data = bsf.load_data(file_name, mode='csv')
distance = 831. # pc
e_distance = 11. # pc
refit = False
run_name = '0'
################################################################################
model_name = 'koester'
limits = {'Te' : [5000, 80000],
'logg' : [ 6.5, 9.5]}
model = bsf.Model(model_name, limits=limits)
################################################################################
star = bsf.Star(name=name,
distance=distance,
e_distance=e_distance,
filters_to_drop=[],
wavelength_range=[0, 1_000_000_000],
data=data,
model=model,
r_limits='blackbody',
run_name=run_name)
################################################################################
star.fit_chi2(refit=refit)
star.fit_noisy_chi2(refit=refit)
################################################################################
star.plot(add_noisy_seds=False,
folder='plots/',
FR_cutoff=0.5)
star.save_summary()
22:39:56 ----- WARNING ----- calculate_chi2
Give "refit=True" if you want to rerun the fitting process.
22:39:56 ----- WARNING ----- get_parameters_from_chi2_minimization
Based on chi2, I recommend removal of following filters: ['WISE/WISE.W1']; chi2=[1722.30668168]
22:39:56 ----- WARNING ----- calculate_noisy_chi2
Give "refit=True" if you want to rerun the fitting process.
22:39:56 ----- WARNING ----- get_realistic_errors_from_iterations
logg_A : The best fit value is at upper limit of the model.
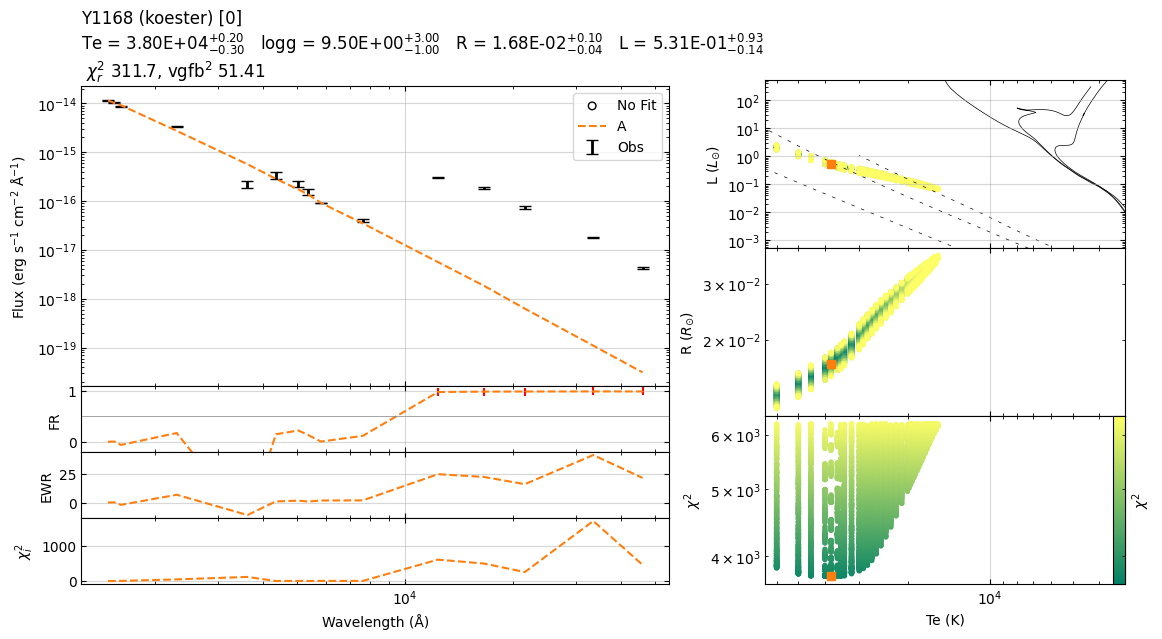
Example 3¶
import src.binary_sed_fitting as bsf
import warnings
warnings.filterwarnings("ignore")
bsf.console.setLevel(bsf.logging.ERROR)
################################################################################
name = 'WOCS2002'
file_name = 'data/extinction_corrected_flux_files/%s.csv'%name
data = bsf.load_data(file_name, mode='csv')
distance = 831. # pc
e_distance = 11. # pc
refit = False
run_name = 'uvblue'
################################################################################
model_name = 'kurucz_uvblue'
limits = {'Te' : [3500, 9000],
'logg' : [ 3, 5],
'MH' : [ 0.0, 0.0]}
model = bsf.Model(model_name, limits=limits)
################################################################################
star = bsf.Star(name=name,
distance=distance,
e_distance=e_distance,
filters_to_drop=[],
wavelength_range=[3000, 1_000_000_000],
data=data,
model=model,
r_limits=[0.1, 10],
run_name=run_name)
################################################################################
star.fit_chi2(refit=refit)
star.fit_noisy_chi2(refit=refit)
################################################################################
star.plot(add_noisy_seds=False,
folder='plots/',
FR_cutoff=0.5)
star.save_summary()
Advance options¶
Console logging options can be changes based on requirements
For minimal logs:
bsf.console.setLevel(bsf.logging.ERROR)For typical logs:
bsf.console.setLevel(bsf.logging.WARNING)For detailed logs:
bsf.console.setLevel(bsf.logging.INFO)For debug logs:
bsf.console.setLevel(bsf.logging.DEBUG)
Editing the
bsffile and re-importing the filepython import importlib importlib.reload(bsf)Starcomponent: Name of the component. Useful for binary fits.
fit_blackbodyFits a blackbody to given flux
p0: Initial guess for the temperature and log(scaling factor). Default is [5000., -20]
fit_chi2_trim: Trimming the number of chi2 fits to save memory
fit_noisy_chi2total_iterations: Number of iterations for noisy fits
plotshow_plot: Showing/hiding the plots in the notebook
plot_publicDuplicate and modify the funcion to suit your need
create_sf_listLOG_SF_STEPSIZE(=0.01) andLOG_SF_FLEXIBILITY(=2) are used to determine the stepsizes in scaling factor
dataDataframe containing the observational data. It is cropped to the fitted filters based on
filters_to_dropandwavelength_range.
data_allA copy of the original data with an added ‘fitted’ column indicating whether the filter is used. Mmodel flux, residuals, and statistical measures are added in
Fitter.get_parameters_from_chi2_minimizationandFitter.get_parameters_from_noisy_chi2_minimization.
data_not_fittedDataFrame containing only removed filters based on
filters_to_dropandwavelength_range
save_figEdit the function to change the plot format (e.g. PDF, PNG or multiple formats)
Star.df_chi2andStar.df_chi2_noisyThe chi2 dataframes are saved in
outputs\.
log files
The raw log files (with logging level DEBUG) are saved in
data\log_<today's date>.txtThe summary logs are chi2 dataframe is saved in
outputs\log_single_fitting.csvandoutputs\log_starsystem_fitting.csv.
import src.binary_sed_fitting as bsf
import warnings
import importlib
warnings.filterwarnings("ignore")
importlib.reload(bsf)
bsf.console.setLevel(bsf.logging.INFO)
################################################################################
name = 'WOCS2002'
file_name = 'data/extinction_corrected_flux_files/%s.csv'%name
data = bsf.load_data(file_name, mode='csv')
distance = 831. # pc
e_distance = 11. # pc
refit = False
################################################################################
model_name = 'kurucz'
limits = {'Te' : [3500, 9000],
'logg' : [ 3, 5],
'MH' : [ 0.0, 0.0],
'alpha': [ 0.0, 0.0]}
model = bsf.Model(model_name, limits=limits)
################################################################################
star = bsf.Star(name=name,
distance=distance,
e_distance=e_distance,
filters_to_drop=['KPNO/Mosaic.I'],
wavelength_range=[3000, 1_000_000_000],
data=data,
model=model,
r_limits='blackbody',
run_name='kur',
component='A')
################################################################################
star.fit_blackbody(p0=[5000., -20],
plot=True,
show_plot=True,
folder=None)
################################################################################
star.fit_chi2(refit=refit,
_trim=1000)
star.fit_noisy_chi2(refit=refit,
total_iterations=100)
################################################################################
star.plot(add_noisy_seds=False,
show_plot=True,
folder=None,
FR_cutoff=0.5)
star.plot_public(add_noisy_seds=False,
show_plot=True,
folder=None,
FR_cutoff=0.5)
22:39:59 ----- INFO ----- __init__
==========================================================
----------------------------------------------------------
------------ WOCS2002 A ------------
----------------------------------------------------------
==========================================================
22:39:59 ----- INFO ----- drop_filters
Fitted Not fitted
wavelength
1481.000000 Astrosat/UVIT.F148W
1541.000000 Astrosat/UVIT.F154W
1608.000000 Astrosat/UVIT.F169M
2303.366368 GALEX/GALEX.NUV
4357.276538 KPNO/Mosaic.B
5035.750275 GAIA/GAIA3.Gbp
5366.240786 KPNO/Mosaic.V
5822.388714 GAIA/GAIA3.G
7619.959993 GAIA/GAIA3.Grp
8101.609574 KPNO/Mosaic.I
8578.159519 GAIA/GAIA3.Grvs
12350.000000 2MASS/2MASS.J
16620.000000 2MASS/2MASS.H
21590.000000 2MASS/2MASS.Ks
33526.000000 WISE/WISE.W1
46028.000000 WISE/WISE.W2
115608.000000 WISE/WISE.W3
22:39:59 ----- INFO ----- drop_filters
Filters: used/all = 12/17
22:39:59 ----- INFO ----- blackbody
Fit parameters: T=5141 K, log_sf=-19.68
22:39:59 ----- INFO ----- blackbody
Fit parameters: T=5141 K, log_sf=-19.68
22:39:59 ----- WARNING ----- calculate_chi2
Give "refit=True" if you want to rerun the fitting process.
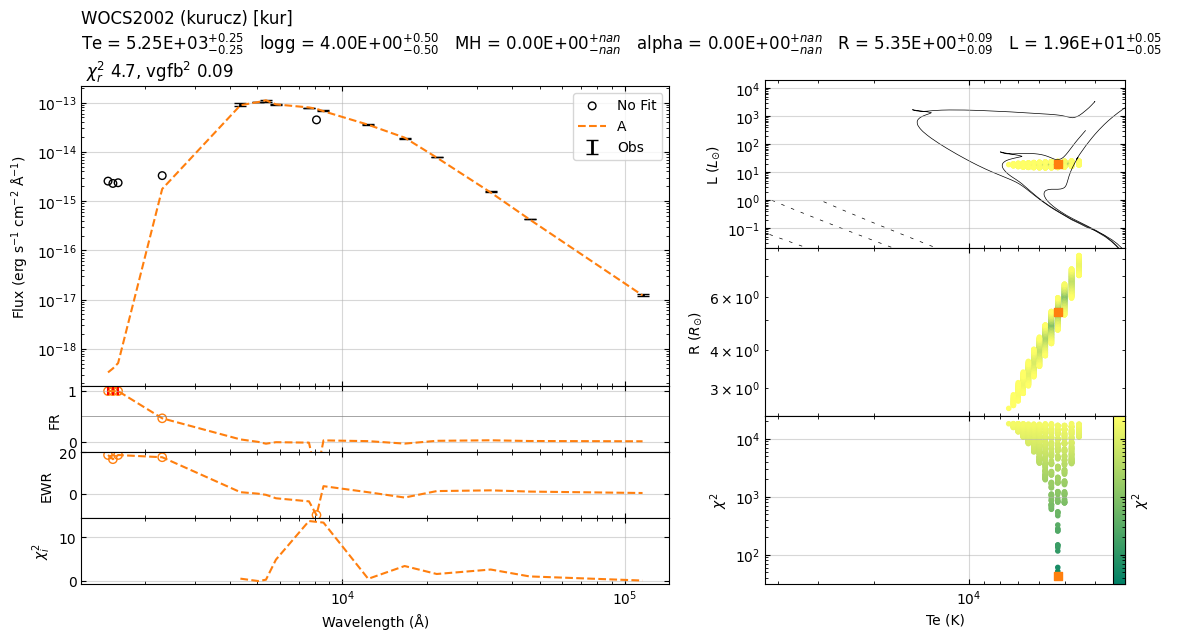
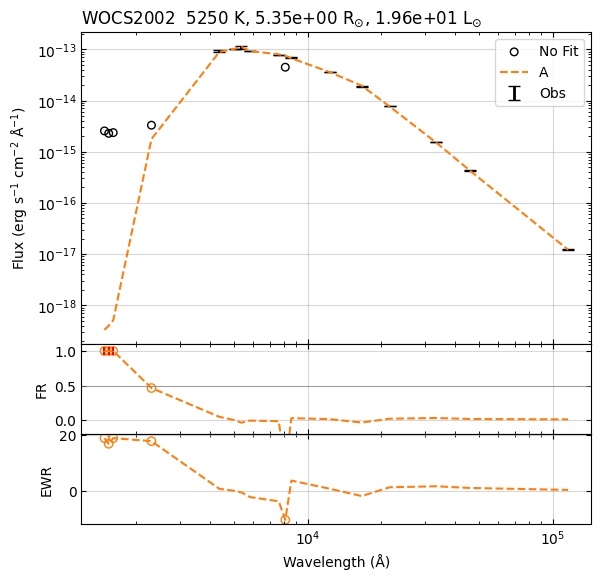
22:39:59 ----- INFO ----- calculate_chi2
Te logg MH alpha sf chi2 R L
0 5250 4.0 0.0 0.0 2.105389e-20 42.148650 5.348053 19.575928
1 5250 4.5 0.0 0.0 2.105389e-20 43.750774 5.348053 19.575928
2 5250 3.5 0.0 0.0 2.105389e-20 46.564792 5.348053 19.575928
3 5250 5.0 0.0 0.0 2.105389e-20 51.654370 5.348053 19.575928
4 5250 3.0 0.0 0.0 2.105389e-20 60.201435 5.348053 19.575928
22:39:59 ----- INFO ----- get_parameters_from_chi2_minimization
Te 5250
logg 4.0
MH 0.0
alpha 0.0
sf 2.1053889442869984e-20
chi2 42.148649603337915
R 5.348053395835723
L 19.57592810070829
vgf2 11.474651537318044
vgfb2 0.8009328529123378
22:39:59 ----- WARNING ----- calculate_noisy_chi2
Give "refit=True" if you want to rerun the fitting process.
22:39:59 ----- INFO ----- calculate_noisy_chi2
Te logg MH alpha sf chi2 R L
0 5250 4.0 0.0 0.0 2.105389e-20 42.148650 5.348053 19.575928
1 5250 4.0 0.0 0.0 2.105389e-20 48.786377 5.348053 19.575928
2 5250 4.5 0.0 0.0 2.105389e-20 58.784976 5.348053 19.575928
3 5250 4.0 0.0 0.0 2.105389e-20 83.279210 5.348053 19.575928
4 5250 4.0 0.0 0.0 2.105389e-20 57.314788 5.348053 19.575928
22:39:59 ----- INFO ----- get_parameters_from_noisy_chi2_minimization
Te 5250(-250,+250)
logg 4.0(-0.5,+0.5)
MH 0.0(-nan,+nan)
alpha 0.0(-nan,+nan)
sf 2.1053889442869984e-20(-4.792450334089126e-22,+4.904080842726991e-22)
R 5.348053395835723(-0.09359116353916891,+0.0940563746344943)
L 19.57592810070829(-0.518255617587945,+0.518255617587945)
print(star.data.index)
print(star.data_all.index)
print(star.data_not_fitted.index)
star.data.head()
Index(['KPNO/Mosaic.B', 'GAIA/GAIA3.Gbp', 'KPNO/Mosaic.V', 'GAIA/GAIA3.G',
'GAIA/GAIA3.Grp', 'GAIA/GAIA3.Grvs', '2MASS/2MASS.J', '2MASS/2MASS.H',
'2MASS/2MASS.Ks', 'WISE/WISE.W1', 'WISE/WISE.W2', 'WISE/WISE.W3'],
dtype='object', name='FilterID')
Index(['Astrosat/UVIT.F148W', 'Astrosat/UVIT.F154W', 'Astrosat/UVIT.F169M',
'GALEX/GALEX.NUV', 'KPNO/Mosaic.B', 'GAIA/GAIA3.Gbp', 'KPNO/Mosaic.V',
'GAIA/GAIA3.G', 'GAIA/GAIA3.Grp', 'KPNO/Mosaic.I', 'GAIA/GAIA3.Grvs',
'2MASS/2MASS.J', '2MASS/2MASS.H', '2MASS/2MASS.Ks', 'WISE/WISE.W1',
'WISE/WISE.W2', 'WISE/WISE.W3'],
dtype='object', name='FilterID')
Index(['Astrosat/UVIT.F148W', 'Astrosat/UVIT.F154W', 'Astrosat/UVIT.F169M',
'GALEX/GALEX.NUV', 'KPNO/Mosaic.I'],
dtype='object', name='FilterID')
| wavelength | flux | error | error_fraction | error_2percent | error_10percent | log_wavelength | log_flux | e_log_flux | fitted | ... | chi2_i | vgf2_i | vgfb2_i | model_flux_median | residual_flux_median | fractional_residual_median | ewr_median | chi2_i_median | vgf2_i_median | vgfb2_i_median | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FilterID | |||||||||||||||||||||
| KPNO/Mosaic.B | 4357.276538 | 9.309407e-14 | 6.130611e-15 | 0.065854 | 6.130611e-15 | 9.309407e-15 | 3.639215 | -13.031078 | 0.028600 | 1 | ... | 0.545712 | 0.545712 | 2.366611e-01 | 8.856525e-14 | 4.528824e-15 | 0.048648 | 0.738723 | 0.545712 | 0.545712 | 2.366611e-01 |
| GAIA/GAIA3.Gbp | 5035.750275 | 1.028575e-13 | 3.881303e-16 | 0.003773 | 2.057150e-15 | 1.028575e-14 | 3.702064 | -12.987764 | 0.001639 | 1 | ... | 0.000081 | 0.000003 | 1.148069e-07 | 1.028540e-13 | 3.485137e-18 | 0.000034 | 0.008979 | 0.000081 | 0.000003 | 1.148069e-07 |
| KPNO/Mosaic.V | 5366.240786 | 1.079375e-13 | 7.108112e-15 | 0.065854 | 7.108112e-15 | 1.079375e-14 | 3.729670 | -12.966827 | 0.028600 | 1 | ... | 0.298089 | 0.298089 | 1.292734e-01 | 1.118184e-13 | -3.880853e-15 | -0.035955 | -0.545975 | 0.298089 | 0.298089 | 1.292734e-01 |
| GAIA/GAIA3.G | 5822.388714 | 9.174216e-14 | 2.539155e-16 | 0.002768 | 1.834843e-15 | 9.174216e-15 | 3.765101 | -13.037431 | 0.001202 | 1 | ... | 4.897209 | 0.093784 | 3.751362e-03 | 9.230407e-14 | -5.619057e-16 | -0.006125 | -2.212964 | 4.897209 | 0.093784 | 3.751362e-03 |
| GAIA/GAIA3.Grp | 7619.959993 | 7.878734e-14 | 3.154436e-16 | 0.004004 | 1.575747e-15 | 7.878734e-15 | 3.881953 | -13.103544 | 0.001739 | 1 | ... | 13.712759 | 0.549535 | 2.198141e-02 | 7.995545e-14 | -1.168111e-15 | -0.014826 | -3.703074 | 13.712759 | 0.549535 | 2.198141e-02 |
5 rows × 24 columns